Visualize HARP2 L2 aerosol product (FastMAPOL)#
Authors: Meng Gao (NASA, SSAI), Sean Foley (NASA, MSU), Kamal Aryal (UMBC)
Summary#
This notebook explores the HARP2 Level 2 (L2) aerosol product derived from the joint aerosol and surface retrieval algorithm: FastMAPOL (Fast Multi-Angle Polarimetric Ocean and Land algorithm). For more detailed information about the algorithm, please refer to the relevant documentation.
Similar to the SPEXone notebook, we will analyze a scene from the Los Angeles wildfire, which includes both smoke and dust events. The analysis will focus on aerosol optical depth, absorption, and size information.
Note that the HARP2 Level 1 (L1) data is still undergoing calibration improvements, which may affect the quality of the L2 data. Data quality is evaluated using several metrics, which are reviewed at the end of this tutorial.
Learning Objectives#
By the end of this notebook, you will understand:
How to acquire HARP2 L2 data
What aerosol products are available
How to visualize basic aerosol properties
How to evaluate data quality
Contents#
1. Setup#
Begin by importing all of the packages used in this notebook. If your kernel uses an environment defined following the guidance on the tutorials page, then the imports will be successful.
from pathlib import Path
import cartopy.crs as ccrs
import earthaccess
import matplotlib.pyplot as plt
import numpy as np
import requests
import xarray as xr
auth = earthaccess.login(persist=True)
fs = earthaccess.get_fsspec_https_session()
2. Get Level-2 Data#
HARP2 L2 data is currently in the test phase with data only available on OB.DAAC, not on earth data cloud yet. We can use the requests library to download data directly from OB.DAAC. The following command line tool downloads one HARP2 FastMAPOL L2 granule at the time stamp 2025/01/09 20:00:19 UTC.
OB_DAAC_PROVISIONAL = "https://oceandata.sci.gsfc.nasa.gov/cgi/getfile/"
HARP2_L2_MAPOL_FILENAME = "PACE_HARP2.20250109T200019.L2.MAPOL_OCEAN.V3_0.nc"
fs.get(f"{OB_DAAC_PROVISIONAL}/{HARP2_L2_MAPOL_FILENAME}", "data/")
paths = list(Path("data").glob("*.nc"))
paths
[PosixPath('data/PACE_HARP2.20250109T200019.L2.MAPOL_OCEAN.V3_0.nc')]
When HARP2 L2 data in provisional level, it will be available through the earth data cloud tools as for the L1C data.
PACE polarimeter L2 products for both HARP2 and SPEXone include four data groups
geolocation_data
geophysical_data
diagnostic_data
sensor_band_parameters
datatree = xr.open_datatree(paths[0])
datatree
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
*empty*
Attributes: (12/31)
title: PACE HARP2 Level-2 aerosol over ocean data
platform: PACE
instrument: HARP2
startDirection: Ascending
endDirection: Ascending
product_name: PACE_HARP2.20250109T200019.L2.MAPOL_OCEAN.V3_0.nc
... ...
time_coverage_start: 2025-01-09T20:00:19Z
time_coverage_end: 2025-01-09T20:05:19Z
reference: https://doi.org/10.5194/amt-16-5863-2023
comments: Test product, bias/scale correction removed
sun_earth_distance: 0.9833925
day_night_flag: DayHere we merge all the data group together for convenience in data manipulations.
dataset = xr.merge(datatree.to_dict().values())
dataset
<xarray.Dataset> Size: 248MB
Dimensions: (number_of_lines: 395, pixels_per_line: 519,
wavelength_3d: 4, number_of_views: 90,
intensity_bands_per_view: 1,
polarization_bands_per_view: 1)
Coordinates:
* wavelength_3d (wavelength_3d) float64 32B 440.0 550.0 670.0 870.0
Dimensions without coordinates: number_of_lines, pixels_per_line,
number_of_views, intensity_bands_per_view,
polarization_bands_per_view
Data variables: (12/56)
latitude (number_of_lines, pixels_per_line) float32 820kB ...
longitude (number_of_lines, pixels_per_line) float32 820kB ...
alh (number_of_lines, pixels_per_line) float32 820kB ...
wind_speed (number_of_lines, pixels_per_line) float32 820kB ...
chla (number_of_lines, pixels_per_line) float32 820kB ...
vd_mode1 (number_of_lines, pixels_per_line) float32 820kB ...
... ...
njev (number_of_lines, pixels_per_line) float64 2MB ...
quality_flag (number_of_lines, pixels_per_line) float64 2MB ...
ozone (number_of_lines, pixels_per_line) float32 820kB ...
surface_pressure (number_of_lines, pixels_per_line) float32 820kB ...
mask_ref (number_of_lines, pixels_per_line, number_of_views, intensity_bands_per_view) float32 74MB ...
mask_dolp (number_of_lines, pixels_per_line, number_of_views, polarization_bands_per_view) float32 74MB ...
Attributes: (12/31)
title: PACE HARP2 Level-2 aerosol over ocean data
platform: PACE
instrument: HARP2
startDirection: Ascending
endDirection: Ascending
product_name: PACE_HARP2.20250109T200019.L2.MAPOL_OCEAN.V3_0.nc
... ...
time_coverage_start: 2025-01-09T20:00:19Z
time_coverage_end: 2025-01-09T20:05:19Z
reference: https://doi.org/10.5194/amt-16-5863-2023
comments: Test product, bias/scale correction removed
sun_earth_distance: 0.9833925
day_night_flag: Day3. Understanding HARP2 L2 product structure#
The HARP2 FastMAPOL L2 product suite includes a long list of aerosol optical properties for both fine and coarse modes (defined in the same format as SPEXone L2 products):
Aerosol optical depth (aot and aot_fine/coarse)
Aerosol single scattering albedo (ssa and ssa_fine/coarse)
Ångström coefficient (angstrom_440_870 and angstrom_440_670)
Aerosol fine mode optical depth fraction (fmf)
etc
As well as aerosol microphysical properties:
Aerosol effective radius (reff_fine/coarse) and variance (veff_fine/coarse)
Aerosol refractive index: real part (mr and mr_fine/coarse), imaginary part (mi and mi_fine/coarse)
Aerosol spherical fraction (sph and sph_fine/coarse)
Aerosol volume density (vd_fine/coarse)
Aerosol fine mode volume fraction (fvf)
Aerosol layer height (alh)
etc
And a set of other products:
Wind speed (wind_speed)
Chlorophyll-a (chla)
Remote sensing reflectance (Rrs*)
The remote sensing reflectance characterizes ocean-leaving reflectance. It is derived via atmospheric correction based on the retrieved aerosol properties at all HARP2 viewing angles. Therefore, it includes an angle dimension, as in the L1C data.
There are two versions of remote sensing reflectance: Rrs1 (before BRDF correction) and Rrs2 (after BRDF correction). Due to the large size of Rrs1 and Rrs2, they are optional outputs in the standard L2 file. Instead, their angular means and standard deviations are typically included as Rrs1_mean/std and Rrs2_mean/std.
datatree["geophysical_data"]
<xarray.DatasetView> Size: 86MB
Dimensions: (number_of_lines: 395, pixels_per_line: 519,
wavelength_3d: 4)
Dimensions without coordinates: number_of_lines, pixels_per_line, wavelength_3d
Data variables: (12/42)
alh (number_of_lines, pixels_per_line) float32 820kB ...
wind_speed (number_of_lines, pixels_per_line) float32 820kB ...
chla (number_of_lines, pixels_per_line) float32 820kB ...
vd_mode1 (number_of_lines, pixels_per_line) float32 820kB ...
vd_mode2 (number_of_lines, pixels_per_line) float32 820kB ...
vd_mode3 (number_of_lines, pixels_per_line) float32 820kB ...
... ...
Rrs2_mean (number_of_lines, pixels_per_line, wavelength_3d) float32 3MB ...
Rrs2_std (number_of_lines, pixels_per_line, wavelength_3d) float32 3MB ...
Rrs1_model_mean (number_of_lines, pixels_per_line, wavelength_3d) float32 3MB ...
Rrs1_model_std (number_of_lines, pixels_per_line, wavelength_3d) float32 3MB ...
Rrs2_model_mean (number_of_lines, pixels_per_line, wavelength_3d) float32 3MB ...
Rrs2_model_std (number_of_lines, pixels_per_line, wavelength_3d) float32 3MB ...4. Visulize HARP2 L2 aerosol properties#
In this example, we visualize the aerosol properties for a scene during LA wild fire with both smoke and dust events. We read the total aerosol optical depth, single scattering albedo, and fine mode volume fraction as below:
aot = dataset["aot"].values
ssa = dataset["ssa"].values
fvf = dataset["fvf"].values
aot.shape, ssa.shape, fvf.shape
((395, 519, 4), (395, 519, 4), (395, 519))
We also need the spatial and angle dimensions as below:
lat = dataset["latitude"].values
lon = dataset["longitude"].values
plot_range = [lon.min(), lon.max(), lat.min(), lat.max()]
wavelength = dataset["wavelength_3d"].values
print(wavelength)
[440. 550. 670. 870.]
For future L2 product, the wavelength variable will be called simple wavelength, rather than wavelength_3d
def plot_l2_product(
data, plot_range, label, title, vmin, vmax, figsize=(12, 4), cmap="viridis"
):
"""Make map and histogram (default)."""
# Create a figure with two subplots: 1 for map, 1 for histogram
fig = plt.figure(figsize=figsize)
gs = fig.add_gridspec(1, 2, width_ratios=[3, 1], wspace=0.3)
# Map subplot
ax_map = fig.add_subplot(gs[0], projection=ccrs.PlateCarree())
ax_map.set_extent(plot_range, crs=ccrs.PlateCarree())
ax_map.coastlines(resolution="110m", color="black", linewidth=0.8)
ax_map.gridlines(draw_labels=True)
# Assume lon and lat are defined globally or passed in
pm = ax_map.pcolormesh(
lon, lat, data, vmin=vmin, vmax=vmax, transform=ccrs.PlateCarree(), cmap=cmap
)
plt.colorbar(pm, ax=ax_map, orientation="vertical", pad=0.1, label=label)
ax_map.set_title(title, fontsize=12)
# Histogram subplot
ax_hist = fig.add_subplot(gs[1])
flattened_data = data[~np.isnan(data)] # Remove NaNs for histogram
valid_count = np.sum(~np.isnan(flattened_data))
ax_hist.hist(
flattened_data, bins=40, color="gray", range=[vmin, vmax], edgecolor="black"
)
ax_hist.set_xlabel(label)
ax_hist.set_ylabel("Count")
ax_hist.set_title("Histogram: N=" + str(valid_count))
# plt.tight_layout()
plt.show()
wavelength_index = 1
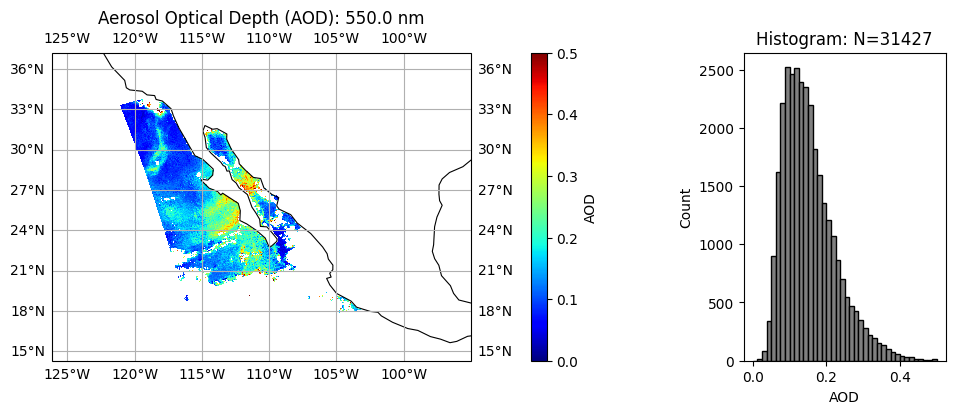
title = "Aerosol Optical Depth (AOD): " + str(wavelength[wavelength_index]) + " nm"
label = "AOD"
data = aot[:, :, wavelength_index]
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=0.5, cmap="jet"
)
wavelength_index = 1
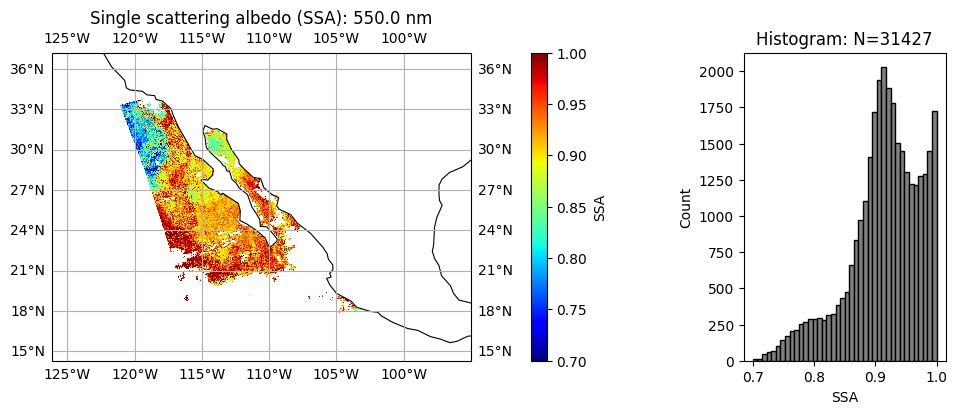
title = "Single scattering albedo (SSA): " + str(wavelength[wavelength_index]) + " nm"
label = "SSA"
data = ssa[:, :, wavelength_index]
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0.7, vmax=1, cmap="jet"
)
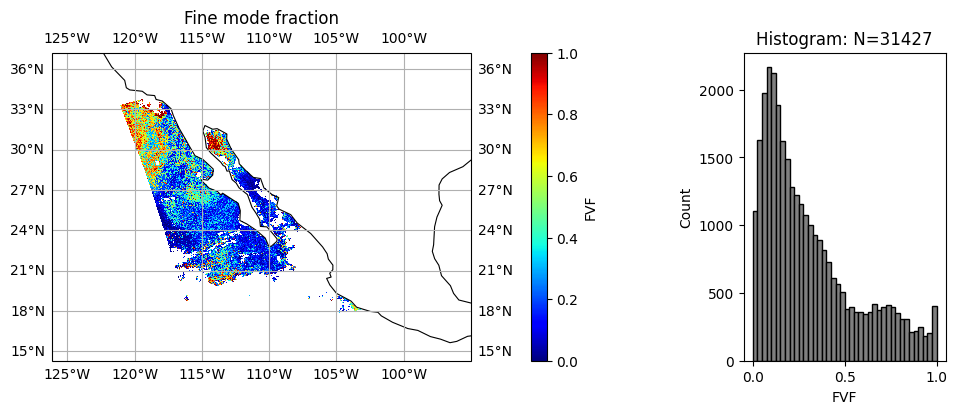
wavelength_index = 1
title = "Fine mode fraction"
label = "FVF"
data = fvf
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=1, cmap="jet"
)
5. Improve data quality: filter low AOD pixels#
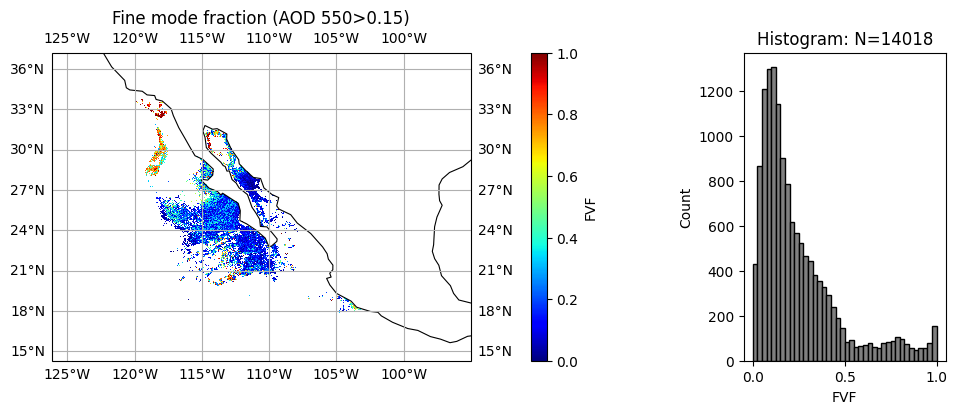
Aerosol absorption and microphysics have larger uncertainties when aerosol loading is low. User can further remove low AOD cases when necessary. We can clearly see, two type of aerosol events with relatively high AOD, the upper one with high absorption (low SSA) and small size (high FVF), probably smoke due to fire; the lower one with less absorption (high SSA) and large size (low FVF), probably dust.
wavelength_index = 1
aot_min = 0.15
title = (
"Filtered single scattering albedo (SSA): "
+ str(wavelength[wavelength_index])
+ " nm (AOD 550>"
+ str(aot_min)
+ ")"
)
label = "SSA"
data = filtered_ssa = np.where(
aot[:, :, wavelength_index] >= aot_min, ssa[:, :, wavelength_index], np.nan
)
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0.7, vmax=1, cmap="jet"
)
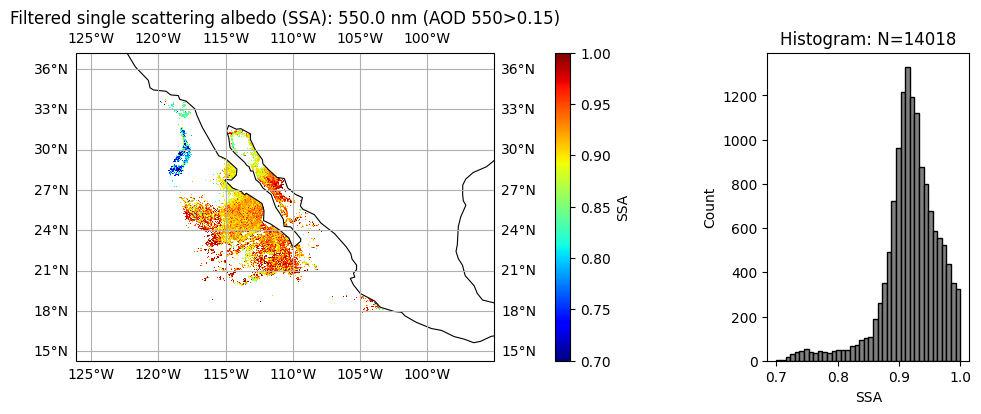
The difference in appearance (after matplotlib automatically normalizes the data) is negligible, but the difference in the physical meaning of the array values is quite important.
wavelength_index = 1
aot_min = 0.15
title = "Fine mode fraction (AOD 550>" + str(aot_min) + ")"
label = "FVF"
data = filtered_ssa = np.where(aot[:, :, wavelength_index] >= aot_min, fvf, np.nan)
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=1, cmap="jet"
)
6. Advanced quality assessment#
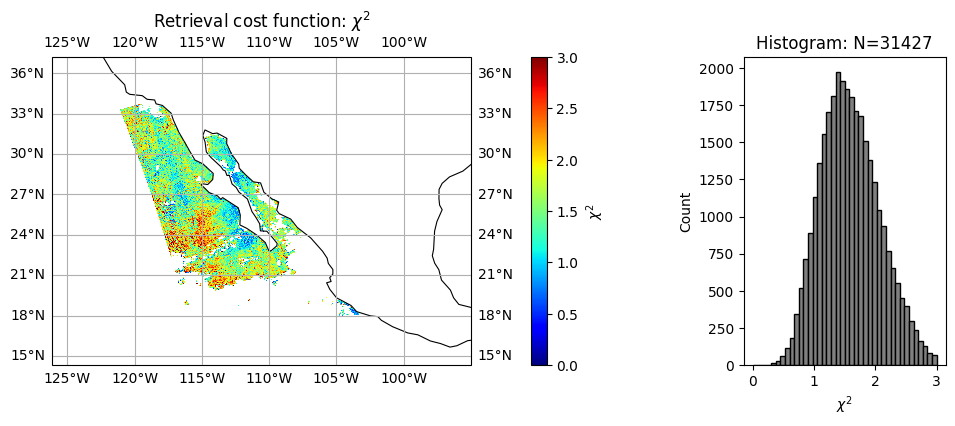
Since the retrieval algorithm is based on optimal estimation by minimizing a \(\chi^2\) cost function defined as the difference between measurement (m) and forward model fitting (f), normalized by total uncertainties (\(\sigma\)).
\(\chi^2 = \frac{1}{N} \sum (f - m)^2/\sigma^2\)
Here N is the total number of measureents used in retreival. The algorithm also adaptively evalue fitting performance, if the fitting perform poor, it will be removed from the retreival process. Therefore, the \(\chi^2\) and \(N\) can be used to evaluate retrieval performance, the pixels with small \(\chi^2\) (good fitting) and large \(N\) (more pixels can be fitted) will better quality. A more quantitatively approach based on error propogation can be also used to compute retrieval uncertainty, which will be include in future product.
To support L3 data processing, a quality flag is also defined, which is usually based on \(\chi^2\) and \(N\). For the HARP2 test data, we choose
quality_flag = 0: when \(\chi^2<1.5\) and \(N_{ref}>60\) and \(N_{DoLP}>60\)
quality_flag = 1: when \(\chi^2<1.5\) and \(N_{ref}>40\) and \(N_{DoLP}>40\)
quality_flag > 1: for higher value \(\chi^2\) and lower values of \(N_{ref}\) and \(N_{DoLP}\) Quality flag will be updated with future L1 data calibration improvement.
chi2 = dataset["chi2"].values
nv_ref = dataset["nv_ref"].values
nv_dolp = dataset["nv_dolp"].values
quality_flag = dataset["quality_flag"].values
title = r"Retrieval cost function: $\chi^2$"
label = r"$\chi^2$"
data = chi2
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=3, cmap="jet"
)
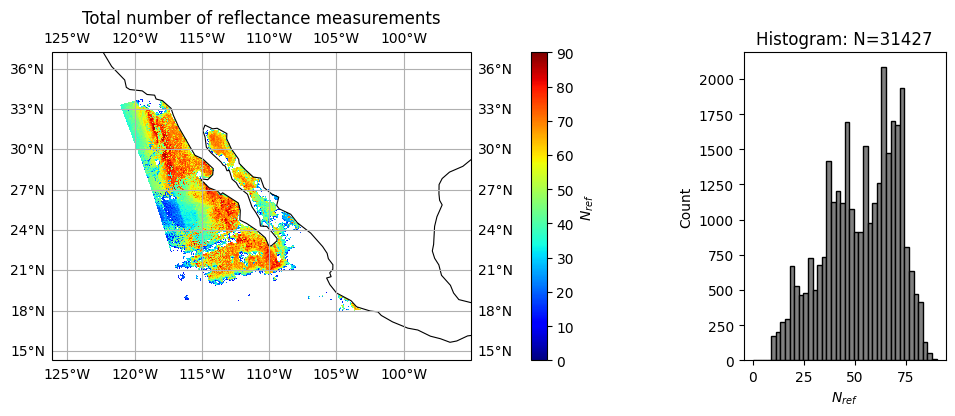
title = r"Total number of reflectance measurements"
label = r"$N_{ref}$"
data = nv_ref
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=90, cmap="jet"
)
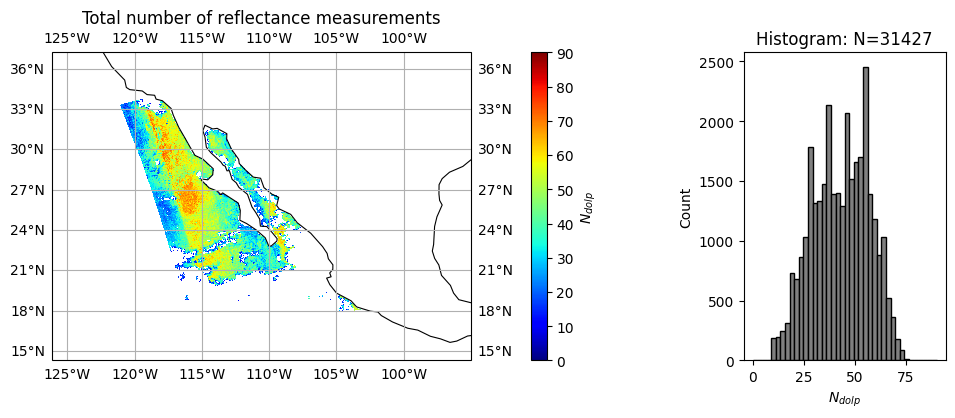
title = r"Total number of reflectance measurements"
label = r"$N_{dolp}$"
data = nv_dolp
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=90, cmap="jet"
)
np.nanmean(chi2), np.nanmean(nv_ref), np.nanmean(nv_dolp)
(np.float32(1.596987),
np.float64(52.5191077735705),
np.float64(42.87914850287969))
Note that \(\chi^2\) converges reasonably well with slight under fitting (averaged around 1.6, peaked around 1.3). Since HARP2 measures 90 angles across 4 bands, the average number of measurement satisfied good fitting are only 52 for reflectance and 42 for polarization, which indicate potential discrepany between forward model and measurements, due to forward model assumptions or likely measurement calibrations.
title = "Retrieval quality flag"
label = "quality_flag"
data = quality_flag
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=4, cmap="cool"
)
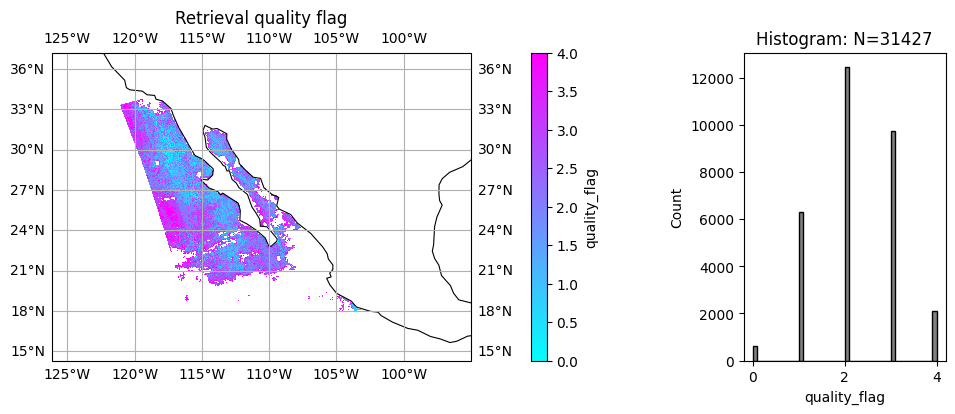
We can evaluate quality flag based on the \(\chi^2\) and \(N\), and only a small portion of data near center of swath reach best quality as we defined as quality_flag=0. With the future improvement of data calibration, more data with better quality will be vailable.
7. Optional: Multi-angle data mask for cloud and data screening#
As mentioned previously, FastMAPOL algorithm conducted internal adaptive data screening on each HARP2 angle, the data mask are provided for both reflectance and DoLP. value 0 means the measurements are used in the retrievals, value 1 or NAN means the measurements are removed from retrieval. Therefore, the adaptive data mask can be also used to evaluate fitting quality and measurement quality at each angle. In the example below, please note the difference pattern for reflectance and polarization, which may indicates different calibration perforance.
mask_ref = dataset["mask_ref"].values
mask_dolp = dataset["mask_dolp"].values
mask_ref.shape, mask_dolp.shape
((395, 519, 90, 1), (395, 519, 90, 1))
angle_index = 5
title = "Adaptive data mask on reflectance: angle index " + str(angle_index)
label = "mask_ref"
data = mask_ref[:, :, angle_index, 0]
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=1, cmap="cool"
)
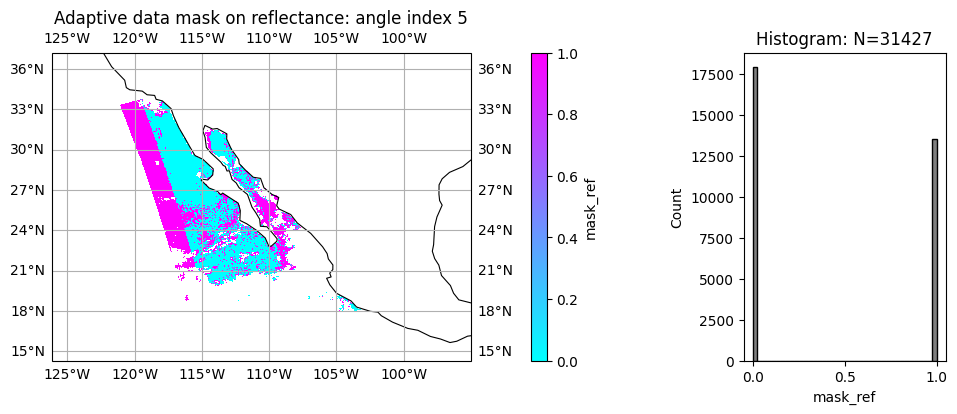
angle_index = 5
title = "Adaptive data mask on DoLP: angle index" + str(angle_index)
label = "mask_DOLP"
data = mask_dolp[:, :, angle_index, 0]
plot_l2_product(
data, plot_range=plot_range, label=label, title=title, vmin=0, vmax=1, cmap="cool"
)
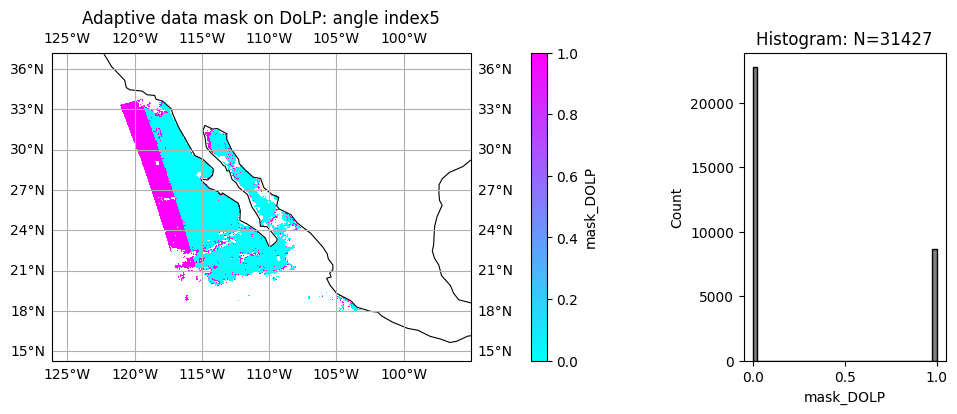
8. Optional: pixel level uncertainty estimation.#
As mentioned previously, pixel level uncertainty can be evalated through error propagation, which propgation measurement uncertainty through Jacobian of the forward model. The estimated uncertainties are discussed in (Gao et al 2021) for HARP2 and AirHARP, but currently not included in the HARP2 L2 products.
9. Reference#
Gao, M., Franz, B. A., Knobelspiesse, K., Zhai, P.-W., Martins, V., Burton, S., Cairns, B., Ferrare, R., Gales, J., Hasekamp, O., Hu, Y., Ibrahim, A., McBride, B., Puthukkudy, A., Werdell, P. J., and Xu, X.: Efficient multi-angle polarimetric inversion of aerosols and ocean color powered by a deep neural network forward model, Atmos. Meas. Tech., 14, 4083–4110, https://doi.org/10.5194/amt-14-4083-2021, 2021.
Gao, M., Knobelspiesse, K., Franz, B. A., Zhai, P.-W., Sayer, A. M., Ibrahim, A., Cairns, B., Hasekamp, O., Hu, Y., Martins, V., Werdell, P. J., and Xu, X.: Effective uncertainty quantification for multi-angle polarimetric aerosol remote sensing over ocean, Atmos. Meas. Tech., 15, 4859–4879, https://doi.org/10.5194/amt-15-4859-2022, 2022.
Gao, M., Franz, B. A., Zhai, P.-W., Knobelspiesse, K., Sayer, A. M., Xu, X., Martins, J. V., Cairns, B., Castellanos, P., Fu, G., Hannadige, N., Hasekamp, O., Hu, Y., Ibrahim, A., Patt, F., Puthukkudy, A., and Werdell, P. J.: Simultaneous retrieval of aerosol and ocean properties from PACE HARP2 with uncertainty assessment using cascading neural network radiative transfer models, Atmos. Meas. Tech., 16, 5863–5881, https://doi.org/10.5194/amt-16-5863-2023, 2023.
