Parallel and Larger-than-Memory Processing#
Authors: Ian Carroll (NASA, UMBC)
The following notebooks are prerequisites for this tutorial.
Learn with OCI: Data Access
Learn with OCI: Processing with Command-line Tools
Learn with OCI: Project and Format
Summary#
Processing a large collection of PACE granules can seem like a big job! The best way to approach a big data processing job is by breaking it into many small jobs, and then putting all the pieces back together again. The reason we break up big jobs has to do with computational resources, specifically memory (i.e. RAM) and processors (i.e. CPUs or GPUs). That large collection of granules can’t all fit in memory; and even if it could, your computation might only use one processor.
This notebook works toward a better understanding of a tool tightly integrated with XArray that breaks up big jobs. The tool is Dask: a Python library for parallel and distributed computing.
Learning Objectives#
At the end of this notebook you will know:
About the framework we’re calling “chunk-apply-combine”
How to start a
daskclient for parallel and larger-than-memory pipelinesOne method for averaging Level-2 “swath” data over time
Contents#
1. Setup#
Begin by importing all of the packages used in this notebook. If your kernel uses an environment defined following the guidance on the tutorials page, then the imports will be successful.
import cartopy.crs as ccrs
import dask.array as da
import earthaccess
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from dask.distributed import Client
from matplotlib.patches import Rectangle
We will discuss dask in more detail below, but we use several additional packages behind-the-scenes. They are installed, but we don’t have to import them directly.
rasteriois a high-level wrapper for GDAL which provides the ability to “warp” with the type of geolocation arrays distributed in Level-2 data.rioxarrayis a wrapper that attaches therasteriotools to XArray data structures
SatPy is another Python package that could be useful for the processing demonstrated in this notebok, especially through its Pyresample toolking. SatPy requires dedicated readers for any given instrument, however, and we have not tested the SatPy reader contributed for PACE/OCI.
The data used in the demonstration is the chlor_a product found in the BGC suite of Level-2 ocean color products from OCI.
bbox = (-77, 36, -73, 41)
results = earthaccess.search_data(
short_name="PACE_OCI_L2_BGC",
temporal=("2024-08-01", "2024-08-07"),
bounding_box=bbox,
)
len(results)
16
The search results include all granules from launch through July of 2024 that intersect a bounding box around the Chesapeake and Delaware Bays. The region is much smaller than an OCI swath, so we do not use the cloud cover search filter which considers the whole swath.
results[0]
paths = earthaccess.open(results[:1])
datatree = xr.open_datatree(paths[0])
dataset = xr.merge(datatree.to_dict().values())
dataset = dataset.set_coords(("latitude", "longitude"))
chla = dataset["chlor_a"]
chla
<xarray.DataArray 'chlor_a' (number_of_lines: 1710, pixels_per_line: 1272)> Size: 9MB
[2175120 values with dtype=float32]
Coordinates:
longitude (number_of_lines, pixels_per_line) float32 9MB ...
latitude (number_of_lines, pixels_per_line) float32 9MB ...
Dimensions without coordinates: number_of_lines, pixels_per_line
Attributes:
long_name: Chlorophyll Concentration, OCI Algorithm
units: mg m^-3
standard_name: mass_concentration_of_chlorophyll_in_sea_water
valid_min: 0.001
valid_max: 100.0
reference: Hu, C., Lee Z., and Franz, B.A. (2012). Chlorophyll-a alg...As a reminder, the Level-2 data has latitude and longitude arrays that give the geolocation of every pixel. The number_of_line and pixels_per_line dimensions don’t have any meaningful coordinates that would be useful for stacking Level-2 files over time. In a lot of the granules, like the one visualized here, there will be a tiny amount of data within the box. But we don’t want to lose a single pixel (if we can help it)!
fig = plt.figure(figsize=(8, 4))
axes = fig.add_subplot()
chla.plot.pcolormesh(x="longitude", y="latitude", robust=True, ax=axes)
axes.add_patch(
Rectangle(
bbox[:2],
bbox[2] - bbox[0],
bbox[3] - bbox[1],
edgecolor="red",
facecolor="none",
)
)
axes.set_aspect("equal")
plt.show()
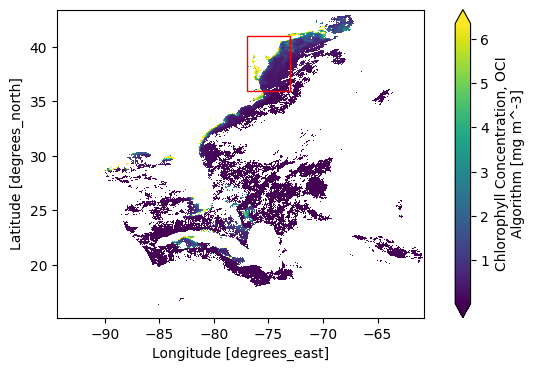
Here is where we first use rasterio and rioxarray.
Those packages together add the rio attribute to the chla dataset which allows us to go GDAL processing in Python.
GDAL requires certain metadata that rio records for us, but it needs to know some information!
The Level-2 file uses the EPSG 4326 coordinate reference system, and the spatial dimensions are:
x (or longitude for EPSG 4326) is “pixels_per_line”
y (or latitude for EPSG 4326) is “number_of_lines”
chla = chla.rio.set_spatial_dims("pixels_per_line", "number_of_lines")
chla = chla.rio.write_crs("epsg:4326")
We can now use the reproject method to grid the L2 data into something like a Level-3 Mapped product, but for a single granule.
chla_L3M = chla.rio.reproject(
dst_crs="epsg:4326",
src_geoloc_array=(
chla.coords["longitude"],
chla.coords["latitude"],
),
)
chla_L3M = chla_L3M.rename({"x":"longitude", "y":"latitude"})
The plotting can now be done with imshow rather than pcolormesh.
plot = chla_L3M.plot.imshow(robust=True)
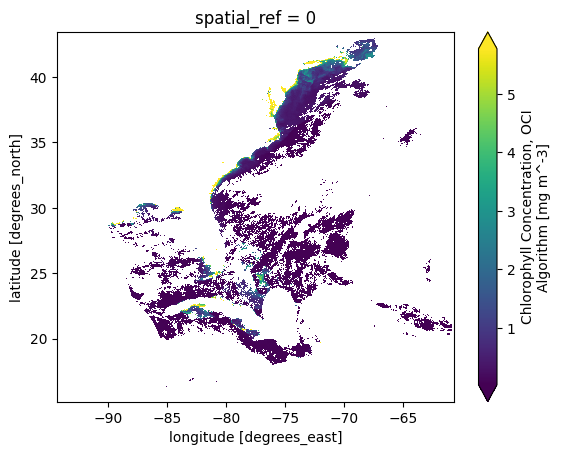
Also, we can easilly select the area-of-interest using our bounding box. This smaller data array will become our “template” for gridding additional granules below.
chla_L3M_aoi = chla_L3M.sel(
{
"longitude": slice(bbox[0], bbox[2]),
"latitude": slice(bbox[3], bbox[1]),
},
)
plot = chla_L3M_aoi.plot.imshow(robust=True)
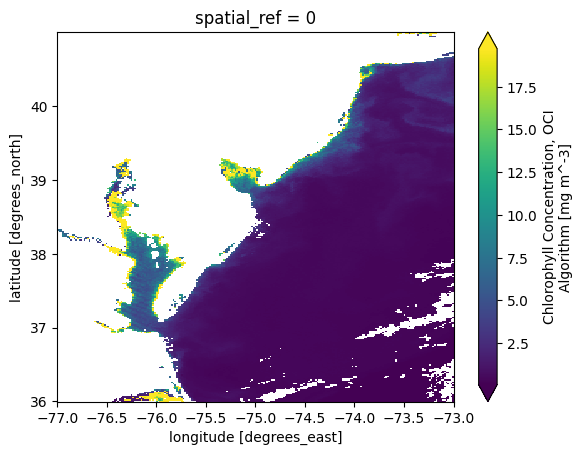
When we get to opening mulitple datasets, we will use a helper function to create a “time” coordinate extracted from metadata.
def time_from_attr(ds):
"""Set the start time attribute as a dataset variable.
Parameters
----------
ds
a dataset corresponding to a Level-2 granule
"""
datetime = ds.attrs["time_coverage_start"].replace("Z", "")
ds["time"] = ((), np.datetime64(datetime, "ns"))
ds = ds.set_coords("time")
return ds
Before we get to data, we will play with some random numbers. Whenever you use random numbers, a good practice is to set a fixed but unique seed, such as the result of secrets.randbits(64).
random = np.random.default_rng(seed=5179916885778238210)
3. Chunk-Apply-Combine#
If you’ve done big data processing, you’ve probably come across the “split-apply-combine” or “map-reduce” frameworks. A simple case is computing group-wise means on a dataset where one variable defines the group and another variable is what you need to average for each group.
split: divide a table into smaller tables, one for each group
apply: calculte the mean on each small table
combine: reassemble the results into a table of group-wise means
The same framework is also used without a natural group by which a dataset should be divided. The split is on equal-sized slices of the original dataset, which we call “chunks”. Rather than a group-wise mean, you could use “chunk-apply-combine” to calculate the grand mean in chunks.
chunk: divide the array into smaller arrays, or “chunks”
apply: calculate the mean and sample size of each chunk (i.e. skipping missing values)
combine: combine the size-weighted means to compute the mean of the whole array
The apply and combine steps have to be capable of calculating results on a slice that can be combined to equal the result you would have gotten on the full array. If a computation can be shoved through “chunk-apply-combine” (see also “map-reduce”), then we can process an array that is too big to read into memory at once. We can also distribute the computation across processors or across a cluster of computers.
We can represent the framework visually using a task graph, a collection of functions (nodes) linked through input and output data (edges).
flowchart LR
A(random.normal) -->|array| B(mean)
The output of the random.normal function becomes the input to the mean function. We can decide to use chunk-apply-combine
if either:
arrayis going to be larger than available memorymeancan be accurately calculated from intermediate results on slices ofarray
By the way, numpy arrays have an nbytes attribute that helps you understand how much memory you may need. Note tha most computations require several times the size of an input array to do the math.
array = random.normal(1, 2, size=2**24)
array
array([-2.21670758, 0.16399032, 2.69695432, ..., 0.95942182,
0.46135808, -0.72953258], shape=(16777216,))
print(f"{array.nbytes / 2**20} MiB")
128.0 MiB
It’s still not too big to fit in memory on most laptops. For demonstration, let’s assume we could fit a 128 MiB array into memory, but not a 1 GiB array. We will calculate the mean of a 1 GiB array anyway, using 8 splits each of size 128 MiB in a serial pipeline.
A simple way to measure performance (i.e. speed) in notebooks is to use the IPython %%timeit magic.
Begin any cell with %%timeit on a line by itself to trigger that cell to run many times under a timer.
How long it takes the cell to run on average will be printed along with any result.
On this system, the serial approach can be seen below to take between 2 and 3 seconds.
%%timeit -r 3
n = 8
s = 0
for _ in range(n):
array = random.normal(1, 2, size=2**24)
s += array.mean()
mean = s / n
3.15 s ± 37.3 ms per loop (mean ± std. dev. of 3 runs, 1 loop each)
All we can implement in a for-loop is serial computation, but we have multiple processors available!
To help visualize the “chunk-apply-combine” framework, we can make a task graph for the “distributed” approach to calculating the mean of a very large array.
%%{ init: { 'flowchart': { 'curve': 'monotoneY' } } }%%
flowchart LR
A(random.normal)
A -->|array_0| C0(apply-mean)
A -->|array_1| C1(apply-mean)
A -->|array_2| C2(apply-mean)
subgraph cluster
C0
C1
C2
end
C0 -->|result_0| D(combine-mean)
C1 -->|result_1| D
C2 -->|result_2| D
In this task graph, the mean function is never used! Instead an apply-mean function and combine-mean functions are used to perform the appropriate computations on chunks and then combine the results to the global mean.
The two things that dask provides is the cluster that distributes jobs for computation as well as many functions (i.e. like apply-mean and combine-mean) that are the “chunk-apply-combine” equivalents of numpy functions.
To begin processing with dask, we start a local client to manage a cluster.
client = Client(n_workers=4, threads_per_worker=1, memory_limit="512MiB")
client
Client
Client-65dc4b21-7221-11f0-8863-86640707dd06
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: /user/itcarroll/proxy/8787/status |
Cluster Info
LocalCluster
bb7ddd98
| Dashboard: /user/itcarroll/proxy/8787/status | Workers: 4 |
| Total threads: 4 | Total memory: 2.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-eb32c1a2-f381-41e4-a92f-ed70c94af3ba
| Comm: tcp://127.0.0.1:42959 | Workers: 0 |
| Dashboard: /user/itcarroll/proxy/8787/status | Total threads: 0 |
| Started: Just now | Total memory: 0 B |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:38191 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/34781/status | Memory: 512.00 MiB |
| Nanny: tcp://127.0.0.1:43369 | |
| Local directory: /tmp/dask-scratch-space/worker-j32driyx | |
Worker: 1
| Comm: tcp://127.0.0.1:44573 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/43543/status | Memory: 512.00 MiB |
| Nanny: tcp://127.0.0.1:43541 | |
| Local directory: /tmp/dask-scratch-space/worker-a1c69w_h | |
Worker: 2
| Comm: tcp://127.0.0.1:46121 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/33783/status | Memory: 512.00 MiB |
| Nanny: tcp://127.0.0.1:41171 | |
| Local directory: /tmp/dask-scratch-space/worker-refakx76 | |
Worker: 3
| Comm: tcp://127.0.0.1:43639 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/34355/status | Memory: 512.00 MiB |
| Nanny: tcp://127.0.0.1:43673 | |
| Local directory: /tmp/dask-scratch-space/worker-_wra1s54 | |
Just like np.random, we can use da.random from dask to generate a data array.
dask_random = da.random.default_rng(random)
dask_array = dask_random.normal(1, 2, size=2**27, chunks=2**22)
dask_array
|
||||||||||||||||
Apply the mean function with out dask_array is quite a bit different.
dask_array.mean()
|
||||||||||||||||
No operation has happened on the array. In fact, the random numbers have not even been generated yet!
No resources are put into action until we explicitly demand a computation; for example, by calling compute, requesting a visualization, or writing data to disk.
%%timeit -r 3
mean = dask_array.mean().compute()
2.02 s ± 64.1 ms per loop (mean ± std. dev. of 3 runs, 1 loop each)
We just demonstrated two ways of doing larger-than-memory calculations.
Our synchronous implemenation (using a for loop) took the strategy of maximizing the use of available memory while processing one chunk: so we used 128 MiB chunks, requiring 8 chunks to get to a 1 GiB array.
Our concurrent implementation (using dask.array), took the strategy of maximizing the use of available processors: so we used small chunks of 32 MiB, requiring 32 to get to a 1 GiB array.
The concurrent implementation was about twice as fast, but your mileage may vary.
client.close()
4. Stacking Level-2 Granules#
The integration of XArray and Dask is designed to let you work without doing much very differently. Of course, there is still a lot of work to do when writing any processing pipeline on a collection of Level-2 granules. Stacking them over time, for instance, may sound easy but requires resampling the data to a common grid.
This section demonstrates one method for stacking Level-2 granules. More details are available in the pre-requiste notebook noted above.
paths = earthaccess.open(results)
paths
[<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240801T174245.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240802T163900.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240802T164400.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240802T181718.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240803T171333.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240803T171833.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240804T161447.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240804T174805.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240805T164420.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240805T164920.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240805T182239.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240806T171853.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240806T172353.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240807T162007.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240807T175325.L2.OC_BGC.V3_0.nc>,
<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20240807T175825.L2.OC_BGC.V3_0.nc>]
kwargs = {"combine": "nested", "concat_dim": "time"}
attrs = xr.open_mfdataset(paths, preprocess=time_from_attr, **kwargs)
attrs
<xarray.Dataset> Size: 128B
Dimensions: (time: 16)
Coordinates:
* time (time) datetime64[ns] 128B 2024-08-01T17:42:45.090000 ... 2024-0...
Data variables:
*empty*
Attributes: (12/45)
title: OCI Level-2 Data BGC
product_name: PACE_OCI.20240801T174245.L2.OC_BGC.V3_...
processing_version: 3.0
history: l2gen par=/data1/sdpsoper/vdc/vpu0/wor...
instrument: OCI
platform: PACE
... ...
geospatial_lon_max: -60.75639
geospatial_lon_min: -94.36226
startDirection: Ascending
endDirection: Ascending
day_night_flag: Day
earth_sun_distance_correction: 0.970880389213562Now, if you try xr.open_mfdataset you will probably encounter an error due to the fact that the number_of_lines is not the same in every granule.
products = xr.open_mfdataset(paths, group="geophysical_data", **kwargs)
---------------------------------------------------------------------------
AlignmentError Traceback (most recent call last)
Cell In[25], line 1
----> 1 products = xr.open_mfdataset(paths, group="geophysical_data", **kwargs)
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/backends/api.py:1706, in open_mfdataset(paths, chunks, concat_dim, compat, preprocess, engine, data_vars, coords, combine, parallel, join, attrs_file, combine_attrs, **kwargs)
1702 try:
1703 if combine == "nested":
1704 # Combined nested list by successive concat and merge operations
1705 # along each dimension, using structure given by "ids"
-> 1706 combined = _nested_combine(
1707 datasets,
1708 concat_dims=concat_dim,
1709 compat=compat,
1710 data_vars=data_vars,
1711 coords=coords,
1712 ids=ids,
1713 join=join,
1714 combine_attrs=combine_attrs,
1715 )
1716 elif combine == "by_coords":
1717 # Redo ordering from coordinates, ignoring how they were ordered
1718 # previously
1719 combined = combine_by_coords(
1720 datasets,
1721 compat=compat,
(...)
1725 combine_attrs=combine_attrs,
1726 )
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/combine.py:367, in _nested_combine(datasets, concat_dims, compat, data_vars, coords, ids, fill_value, join, combine_attrs)
364 _check_shape_tile_ids(combined_ids)
366 # Apply series of concatenate or merge operations along each dimension
--> 367 combined = _combine_nd(
368 combined_ids,
369 concat_dims,
370 compat=compat,
371 data_vars=data_vars,
372 coords=coords,
373 fill_value=fill_value,
374 join=join,
375 combine_attrs=combine_attrs,
376 )
377 return combined
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/combine.py:246, in _combine_nd(combined_ids, concat_dims, data_vars, coords, compat, fill_value, join, combine_attrs)
242 # Each iteration of this loop reduces the length of the tile_ids tuples
243 # by one. It always combines along the first dimension, removing the first
244 # element of the tuple
245 for concat_dim in concat_dims:
--> 246 combined_ids = _combine_all_along_first_dim(
247 combined_ids,
248 dim=concat_dim,
249 data_vars=data_vars,
250 coords=coords,
251 compat=compat,
252 fill_value=fill_value,
253 join=join,
254 combine_attrs=combine_attrs,
255 )
256 (combined_ds,) = combined_ids.values()
257 return combined_ds
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/combine.py:278, in _combine_all_along_first_dim(combined_ids, dim, data_vars, coords, compat, fill_value, join, combine_attrs)
276 combined_ids = dict(sorted(group))
277 datasets = combined_ids.values()
--> 278 new_combined_ids[new_id] = _combine_1d(
279 datasets, dim, compat, data_vars, coords, fill_value, join, combine_attrs
280 )
281 return new_combined_ids
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/combine.py:301, in _combine_1d(datasets, concat_dim, compat, data_vars, coords, fill_value, join, combine_attrs)
299 if concat_dim is not None:
300 try:
--> 301 combined = concat(
302 datasets,
303 dim=concat_dim,
304 data_vars=data_vars,
305 coords=coords,
306 compat=compat,
307 fill_value=fill_value,
308 join=join,
309 combine_attrs=combine_attrs,
310 )
311 except ValueError as err:
312 if "encountered unexpected variable" in str(err):
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/concat.py:277, in concat(objs, dim, data_vars, coords, compat, positions, fill_value, join, combine_attrs, create_index_for_new_dim)
264 return _dataarray_concat(
265 objs,
266 dim=dim,
(...)
274 create_index_for_new_dim=create_index_for_new_dim,
275 )
276 elif isinstance(first_obj, Dataset):
--> 277 return _dataset_concat(
278 objs,
279 dim=dim,
280 data_vars=data_vars,
281 coords=coords,
282 compat=compat,
283 positions=positions,
284 fill_value=fill_value,
285 join=join,
286 combine_attrs=combine_attrs,
287 create_index_for_new_dim=create_index_for_new_dim,
288 )
289 else:
290 raise TypeError(
291 "can only concatenate xarray Dataset and DataArray "
292 f"objects, got {type(first_obj)}"
293 )
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/concat.py:523, in _dataset_concat(datasets, dim, data_vars, coords, compat, positions, fill_value, join, combine_attrs, create_index_for_new_dim)
520 # Make sure we're working on a copy (we'll be loading variables)
521 datasets = [ds.copy() for ds in datasets]
522 datasets = list(
--> 523 align(
524 *datasets, join=join, copy=False, exclude=[dim_name], fill_value=fill_value
525 )
526 )
528 dim_coords, dims_sizes, coord_names, data_names, vars_order = _parse_datasets(
529 datasets
530 )
531 dim_names = set(dim_coords)
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/alignment.py:944, in align(join, copy, indexes, exclude, fill_value, *objects)
748 """
749 Given any number of Dataset and/or DataArray objects, returns new
750 objects with aligned indexes and dimension sizes.
(...)
934
935 """
936 aligner = Aligner(
937 objects,
938 join=join,
(...)
942 fill_value=fill_value,
943 )
--> 944 aligner.align()
945 return aligner.results
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/alignment.py:637, in Aligner.align(self)
635 self.find_matching_unindexed_dims()
636 self.align_indexes()
--> 637 self.assert_unindexed_dim_sizes_equal()
639 if self.join == "override":
640 self.override_indexes()
File /srv/conda/envs/notebook/lib/python3.11/site-packages/xarray/structure/alignment.py:499, in Aligner.assert_unindexed_dim_sizes_equal(self)
497 add_err_msg = ""
498 if len(sizes) > 1:
--> 499 raise AlignmentError(
500 f"cannot reindex or align along dimension {dim!r} "
501 f"because of conflicting dimension sizes: {sizes!r}" + add_err_msg
502 )
AlignmentError: cannot reindex or align along dimension 'number_of_lines' because of conflicting dimension sizes: {1709, 1710}
Even if they were consistent, each granule has a different array of coordinates for latitude and longitude.
Before doing a calculation over time, we need to project the Level-2 arrays to a common grid.
Here is a function to do the same kind of gridding as above, but allowing us to pass the “destination” (.i.e. dst)
projection parameters.
def grid_match(path, dst_crs, dst_shape, dst_transform):
"""Reproject a Level-2 granule to match a Level-3M-ish granule."""
dt = xr.open_datatree(path)
da = dt["geophysical_data"]["chlor_a"]
da = da.rio.set_spatial_dims("pixels_per_line", "number_of_lines")
da = da.rio.set_crs("epsg:4326")
da = da.rio.reproject(
dst_crs,
shape=dst_shape,
transform=dst_transform,
src_geoloc_array=(
dt["navigation_data"]["longitude"],
dt["navigation_data"]["latitude"],
),
)
da = da.rename({"x":"longitude", "y":"latitude"})
return da
Let’s try out the function, using chla_L3M_aoi as our template.
crs = chla_L3M_aoi.rio.crs
shape = chla_L3M_aoi.rio.shape
transform = chla_L3M_aoi.rio.transform()
grid_match(paths[0], crs, shape, transform)
<xarray.DataArray 'chlor_a' (latitude: 296, longitude: 237)> Size: 281kB
array([[ nan, nan, nan, ..., nan,
nan, 1.0616910e+02],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[ nan, nan, nan, ..., nan,
nan, 6.8380810e-02],
[ nan, nan, nan, ..., nan,
nan, 6.6567786e-02],
[ nan, nan, nan, ..., nan,
nan, 6.5402456e-02]], shape=(296, 237), dtype=float32)
Coordinates:
* longitude (longitude) float64 2kB -76.99 -76.98 -76.96 ... -73.02 -73.01
* latitude (latitude) float64 2kB 40.99 40.97 40.96 ... 36.04 36.02 36.01
spatial_ref int64 8B 0
Attributes:
long_name: Chlorophyll Concentration, OCI Algorithm
units: mg m^-3
standard_name: mass_concentration_of_chlorophyll_in_sea_water
valid_min: 0.001
valid_max: 100.0
reference: Hu, C., Lee Z., and Franz, B.A. (2012). Chlorophyll-a alg...Now that we have encapusulated our processing in a function, we can use dask to distribute computation.
As before, we need a client.
client = Client()
client
Client
Client-74a159f1-7221-11f0-8863-86640707dd06
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: /user/itcarroll/proxy/8787/status |
Cluster Info
LocalCluster
3fab91f9
| Dashboard: /user/itcarroll/proxy/8787/status | Workers: 4 |
| Total threads: 4 | Total memory: 3.71 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-1aeb7137-0154-409d-82d3-fdabdc23bb0d
| Comm: tcp://127.0.0.1:38341 | Workers: 0 |
| Dashboard: /user/itcarroll/proxy/8787/status | Total threads: 0 |
| Started: Just now | Total memory: 0 B |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:41187 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/38719/status | Memory: 0.93 GiB |
| Nanny: tcp://127.0.0.1:42775 | |
| Local directory: /tmp/dask-scratch-space/worker-nzqhkop4 | |
Worker: 1
| Comm: tcp://127.0.0.1:41801 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/40667/status | Memory: 0.93 GiB |
| Nanny: tcp://127.0.0.1:41601 | |
| Local directory: /tmp/dask-scratch-space/worker-o879xsw8 | |
Worker: 2
| Comm: tcp://127.0.0.1:35243 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/33035/status | Memory: 0.93 GiB |
| Nanny: tcp://127.0.0.1:44101 | |
| Local directory: /tmp/dask-scratch-space/worker-e9b4q_in | |
Worker: 3
| Comm: tcp://127.0.0.1:37293 | Total threads: 1 |
| Dashboard: /user/itcarroll/proxy/46157/status | Memory: 0.93 GiB |
| Nanny: tcp://127.0.0.1:40615 | |
| Local directory: /tmp/dask-scratch-space/worker-0k17q4p3 | |
Call client.map to send grid_match to the cluster and prepare to run it independently on each elements of paths.
futures = client.map(
grid_match,
paths,
dst_crs=crs,
dst_shape=shape,
dst_transform=transform,
)
futures
[<Future: pending, key: grid_match-ea6f45dda1328be13a653990c0269be2>,
<Future: pending, key: grid_match-0043f538efb6bf582abc5f86f1603750>,
<Future: pending, key: grid_match-b108cd9fc4c0e16d0811c8734f50e71e>,
<Future: pending, key: grid_match-706ca2fb43c1f3a885dfac51397916c2>,
<Future: pending, key: grid_match-7134041f2566f1252e20dc3bb3d6ab43>,
<Future: pending, key: grid_match-58cb9221fe5285a65d90148e16251c37>,
<Future: pending, key: grid_match-02a488c341a138e3395a0c69622dbd16>,
<Future: pending, key: grid_match-0609a2b1be2218dbec3cb35378c38b49>,
<Future: pending, key: grid_match-eab1133578fd3958b15898a3d68857b4>,
<Future: pending, key: grid_match-9b559e4abfbb0fa357edd2a3dabca251>,
<Future: pending, key: grid_match-2b990fd98129a799561f54ad06e7ced6>,
<Future: pending, key: grid_match-e5cd4c1ffb07c3038c48614eec04f4d6>,
<Future: pending, key: grid_match-bd06e29c7221da79764b48ce7e61395a>,
<Future: pending, key: grid_match-7ab512662e9eae710e2186af38b649c4>,
<Future: pending, key: grid_match-be28ed8ebcc9956b072cb8da4e33e1d4>,
<Future: pending, key: grid_match-8a3e23533a455688d82b510065ead46f>]
The futures will remain pending until the tasks have been completed on the cluster.
You don’t need to wait for them; the next call to client.gather will wait for the results.
These results can now be easilly stacked
chla = xr.combine_nested(client.gather(futures), concat_dim="time")
chla["time"] = attrs["time"]
chla
<xarray.DataArray 'chlor_a' (time: 16, latitude: 296, longitude: 237)> Size: 4MB
array([[[ nan, nan, nan, ...,
nan, nan, 1.0616910e+02],
[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
...,
[ nan, nan, nan, ...,
nan, nan, 6.8380810e-02],
[ nan, nan, nan, ...,
nan, nan, 6.6567786e-02],
[ nan, nan, nan, ...,
nan, nan, 6.5402456e-02]],
[[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
...
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan]],
[[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
...,
[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan],
[ nan, nan, nan, ...,
nan, nan, nan]]],
shape=(16, 296, 237), dtype=float32)
Coordinates:
* longitude (longitude) float64 2kB -76.99 -76.98 -76.96 ... -73.02 -73.01
* latitude (latitude) float64 2kB 40.99 40.97 40.96 ... 36.04 36.02 36.01
spatial_ref int64 8B 0
* time (time) datetime64[ns] 128B 2024-08-01T17:42:45.090000 ... 20...plot = chla.mean("time").plot.imshow(robust=True)
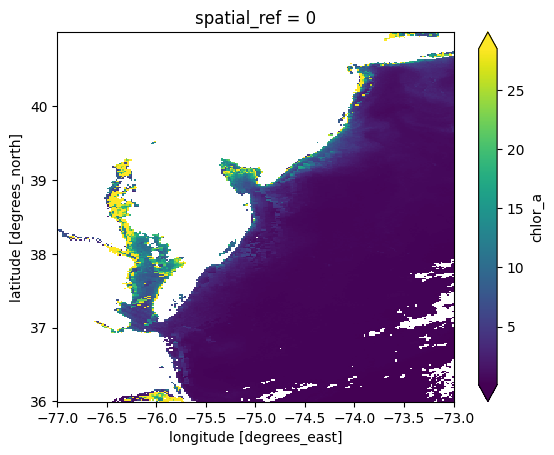
client.close()
5. Scaling Out#
The example above relies on a LocalCluster, which only uses the resources on the JupyterLab server running your notebook.
The promise of the commercial cloud is massive scalability.
Again, dask and the CryoCloud come to our aid in the form of a pre-configured dask_gateway.Gateway.
The gateway object created below works with the CryoCloud to launch additional servers,
and now those servers are enlisted in your cluster.
from dask_gateway import Gateway
gateway = Gateway()
options = gateway.cluster_options()
options
cluster = gateway.new_cluster(options)
cluster
The cluster starts with no workers, and you must set manual or adaptive scaling in order to get any workers.
The cluster can take several minutes to spin up. Monitor the dashboard to ensure you have workers.
client = cluster.get_client()
client
Client
Client-97f811f9-7221-11f0-8863-86640707dd06
| Connection method: Cluster object | Cluster type: dask_gateway.GatewayCluster |
| Dashboard: /services/dask-gateway/clusters/prod.899a01c54ac14b97acc2bbc1f5b6d35b/status |
Cluster Info
GatewayCluster
- Name: prod.899a01c54ac14b97acc2bbc1f5b6d35b
- Dashboard: /services/dask-gateway/clusters/prod.899a01c54ac14b97acc2bbc1f5b6d35b/status
Use this client exactly as above:
futures = client.map(
grid_match,
paths,
dst_crs=crs,
dst_shape=shape,
dst_transform=transform,
)
chla = xr.combine_nested(client.gather(futures), concat_dim="time")
chla["time"] = attrs["time"]
chla
cluster.close()

